
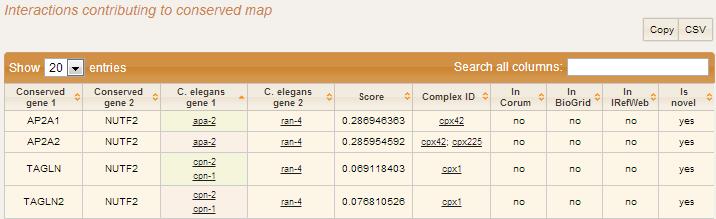
Is mapped to the degree of the node (number of edges connected to Using thresholds set for the gal1RGexp experiment bundled withĬytoscape in the sampleData/galExpData.csv file). Low expression ratio and yellow represents a high expression ratio, Gene expression values for each node are colored alongĪ color gradient between blue and yellow (where blue represents a
Or for nodes/edges not covered by a mapping for a particular property. The Default Value is used when no mapping is defined for a property, There is no Bypass option for Node Table styles. Clicking on theīypass column for selected node(s)/edge(s) allows you toĮnter a bypass for that property for selected node(s)/edge(s). Note that a node/edge or subset of nodes/edges must be Bypass displays any style bypass for a selected node.Property expands the property entry to show the interface forĮditing the mapping. Mapping displays the type of mapping currently in use for.Property allows you to change the default value. Clicking on the Default Value column for any The Default Value shows just that, the default value for.Each property entry in the list has 3 columns:.At the top of the list a Properties drop-downĪllows you to add additional properties to the list. Each tab contains a list of properties relevant to theĬurrent style.Note that style selections for the Table Panel are not saved as part of the current style. The main area of the interface is composed of four tabs, for Node,Įdge and Network properties, as well as a Table tab for style options for the Table Panel, for setting mappings on individual columns in the Node and Edge tables.Options to rename, remove, create and copy a Style, and an option to This will reveal a set of buttons for selecting and deleting styles. The list of available styles can be edited by clicking the Edit.The Show only Applied Styles button will filter the list to display only the applied style.The list of available styles is searchable using the search field.At the top of the interface, there is a drop-down for selecting.With a specific style selected, the Style panel displays theĭetails for a given style and is used to edit these details as well. Styles using the drop-down and the Options menu. This interface allows you to create/delete/view and switch between different
The Style interface is located under the Style panel of the Show highly-connected region by edge bundling and opacity.Īdd photo/image/graphics on top of nodes.Ĭytoscape 3 has several sample styles. Use specific line types to indicate different types of interactions.Ĭontrol edge transparency (opacity) using edge weights.Ĭontrol multiple edge properties using edge score.īrowse extremely-dense networks by controlling the opacity of nodes. Visualize gene expression data along a color gradient.Įncode specific physical entities as different node shapes. …or, set the font size of the node labels instead. Set node sizes based on the degree of connectivity of the nodes. Specify a default color and shape for all nodes. Interface, the appearance of your network is easily customized. Mapped table data sets is called a Style and can be created orĮdited in the Style panel of the Control Panel. Transparency, or font type) of the network. One of Cytoscape’s strengths in network visualization is the ability toĪllow users to encode any table data (name, type, degree, weight,Įxpression data, etc.) as a property (such as color, size of node,


 0 kommentar(er)
0 kommentar(er)
